About Dynamic Code,
Dynamic Code is a HealthTech company enabling remote diagnostics through tests with self-sampling and laboratory analysis. The concept is offered both through digital healthcare partners and directly to the consumer through a platform which combines a physical test with a digital customer journey. Through DNA-technique and a digital full-service solution the customer receives an analytical test result and healthcare follow-up in a safe an efficient way.
The passion & purpose of Dynamic Code is “Shaping tomorrow’s diagnostics – enabling more people to live a healthier life”. In order to work towards this, quality management needs to be an integral part of everything we do. Quality and sustainability is also one of our four cornerstones, representing “what” we do to achieve our targets. As proud professionals we always work in accordance with applicable laws, regulations, standards, and other requirements set in our business, which means that we must follow our quality management system based on ISO13485:2016 and ISO17025. Furthermore, it is of great importance to Dynamic Code to act according to IVDR 2017/746 and other applicable laws and standards.
Together we build our future and through uniform and efficient working methods and processes, a common understanding of roles and responsibilities. A desire for continuous improvement throughout the business we contribute to achieving our goals and meeting our own, and our customers' quality requirements.
At Dynamic Code we aim higher and all employees of Dynamic Code are committed to consistent provision of quality products and services that will satisfy the demands and expectations of customers by continual improvement of the quality management system and conforming to local standards and prevailing codes of practice.
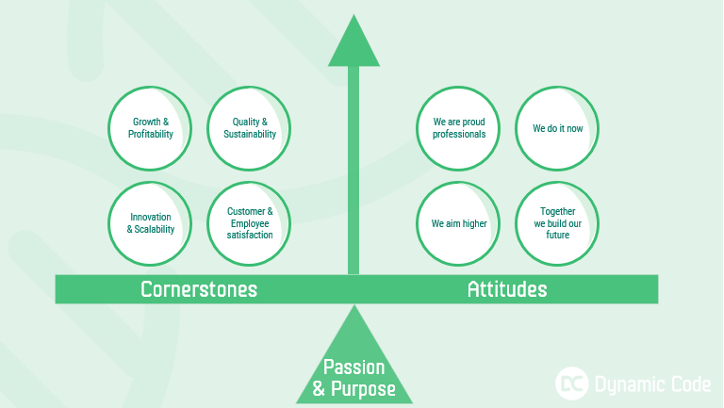
To ensure adherence to our quality policy, all employees must ensure the below:
- To have good knowledge of and apply applicable laws and regulations that affect our business
- To meet our customers' expectations through high delivery precision and competent execution that applies to each customer
- To develop and perform safe analysis
- To ensure impartiality and no conflict of interest
- To carry out our work with commitment, with the right competence and with good routines that are set in accordance with current rules and regulations
- To work systematically with self-control and experience feedback as a method for achieving continuous improvements of our routines, templates, and reference documents
- To give a good impression and trust and work for a healthy work environment
- To cooperate and work without prestige. Our goal is to have good relations with other players in the healthcare chain
- To have active, responsive, and developing leadership
- To be transparent with our quality results and constantly work on improvements
The quality policy covers all operations within Dynamic Code. All employees and those who work for Dynamic Code have a responsibility to follow this quality policy.